tl;dr as @HongOoi is telling you, wait and Adhesive are confounded in your model. lme is a little stupider/more stubborn than many of the other modeling functions in R, which will either warn you explicitly that you have confounded fixed effects or automatically drop some of them for you.
It’s a bit easier to see this if you plot the data:
## source("SO50505290_data.txt")
library(ggplot2)
ggplot(dd,aes(Adhesive,data_mTBS,
fill=Aging,
alpha=Approach))+
facet_grid(.~wait,scale="free_x",space="free",
labeller=label_both)+
guides(alpha = guide_legend(override.aes = list(fill = "darkgray")))+
geom_boxplot()
ggsave("SO50505290.png")
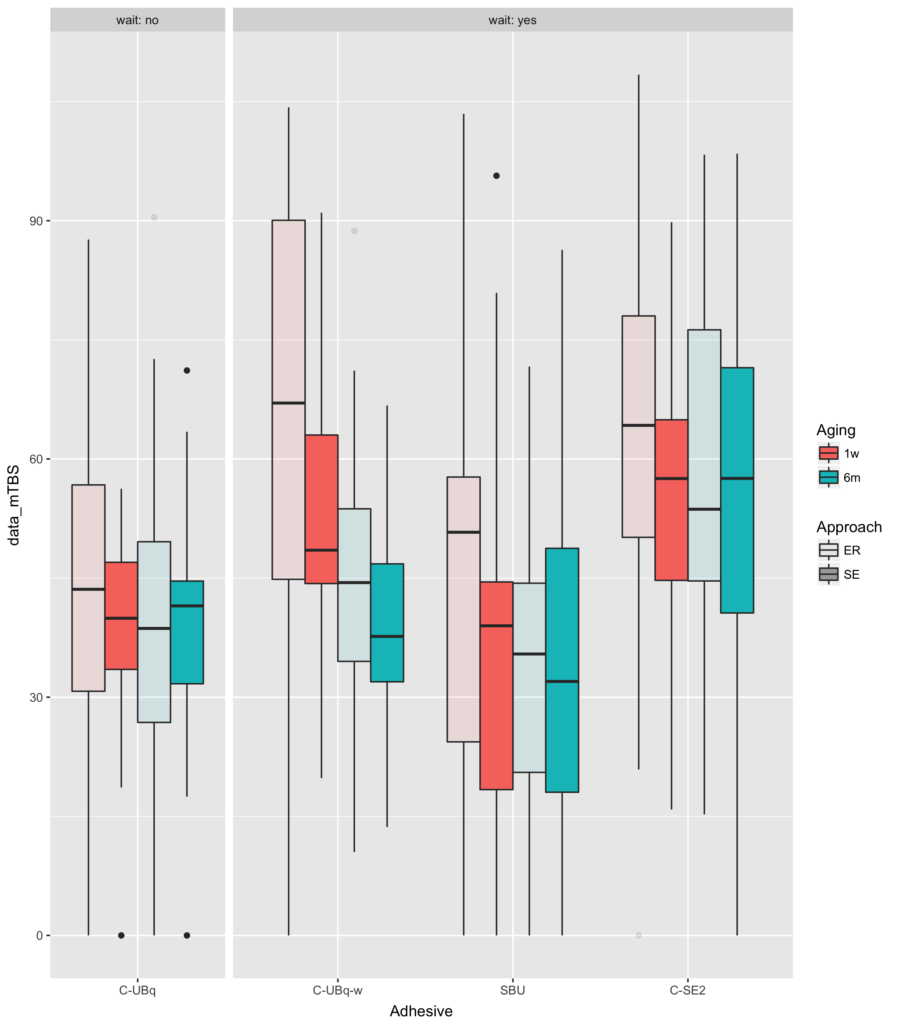
This shows you that knowing that wait=="no" is the same as knowing that Adhesive=="C-UBq".
It would probably make more sense to back up and think about the questions you’re asking, but if you do this with lme4::lmer it will tell you
fixed-effect model matrix is rank deficient so dropping 16 columns / coefficients
library(lme4)
LME_02<-lmer(data_mTBS ~ Adhesive*Approach*Aging*wait+
(1|data_name),
na.action=na.exclude,data = dd)
